ABOUT YODEAH

OUR STORY
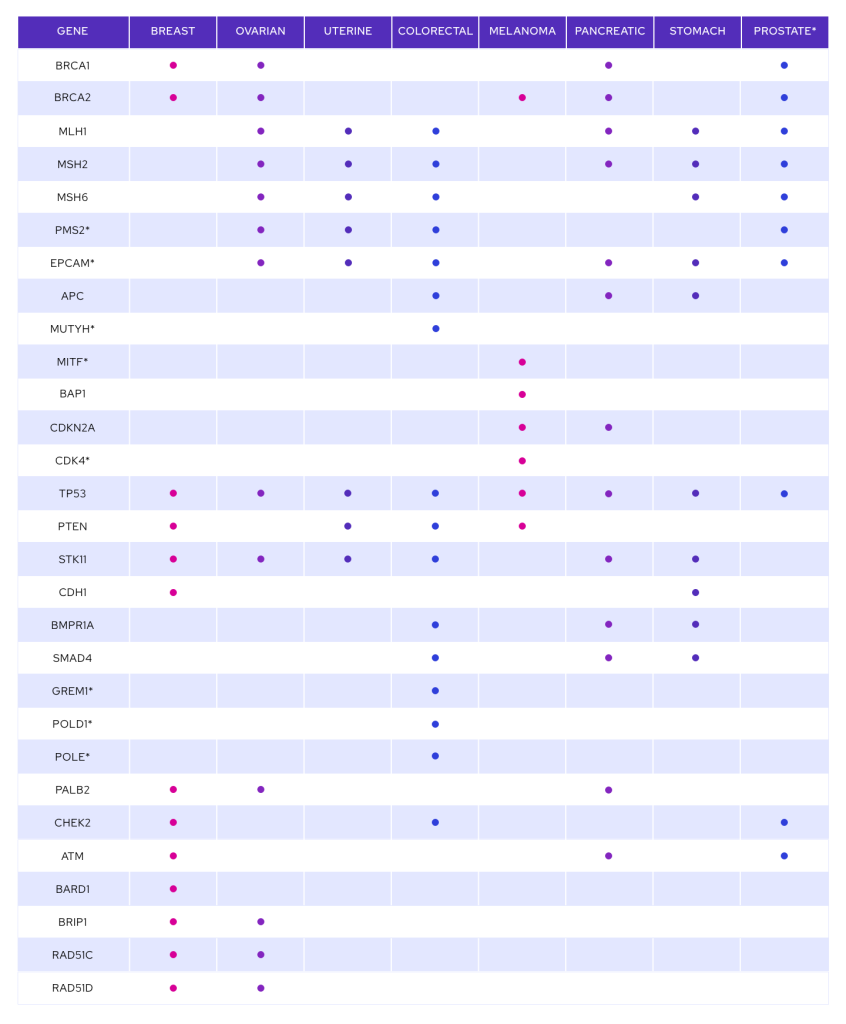
Ashkenazi Jewish men and women are more likely to carry hereditary genetic mutations that can significantly increase their lifetime risk of cancer. Most people do not KNOW and will not have a family history. Yodeah’s mission is to educate the Jewish community about hereditary cancer genetic mutations and provide access to affordable, clinical-grade testing. Yodeah’s goal is to save lives.
Knowledge is power.
ELIZABETH ETKIN-KRAMER, M.D, F.A.C.O.G, FOUNDER
Dr. Etkin-Kramer has been a board-certified obstetrician/gynecologist in Miami Beach since 1993. She holds teaching positions at both FIU and University of Miami medical schools. She has a particular interest in breast and ovarian cancer risk assessment and has educated other healthcare providers on risk assessment inclusive genetic testing. As the scientific literature over the years has clearly shown that Ashkenazi Jews are more likely to inherit BRCA mutations regardless of family history, and most are unaware, she has been a vocal proponent of population-based genetic testing in the Ashkenazi Jewish community. She co-founded the Prevent Hereditary Cancer Coalition with Lauren Corduck (z”l) to identify carriers to mitigate hereditary cancer burden in those individuals and families.

OUR VOLUNTEERS
BOARD MEMBERS



Lauren Harshman Davis, M.D.

Rabbi Laila Haas

Adrienne Pardo, Esq.


Gina Russ, Esq.

Robin Strauss Furlong, M.D.

MEDICAL ADVISORY BOARD
Line Bohbot,
PA-C
Cancer Genetic Counselor
MD MS
M.D.
M.D.
RABBINIC ADVISORY BOARD
(Chair)
THANK YOU TO OUR SPONSORS FOR SUPPORTING YODEAH’S MISSION